|
| 1 | +# GARI |
| 2 | +GARI (Genetic Algorithm for Reproducing Images) is a Python project that uses the PyGAD library for reproducing images using the genetic algorithm. GARI reproduces a single image using Genetic Algorithm (GA) by evolving pixel values. This project works with both color and gray images. |
| 3 | + |
| 4 | +For implementing the genetic algorithm, the PyGAD library is used. Check its documentation here: https://pygad.readthedocs.io |
| 5 | + |
| 6 | +**IMPORTANT** *If you are coming for the code of the tutorial [Reproducing Images using a Genetic Algorithm with Python](https://heartbeat.fritz.ai/reproducing-images-using-a-genetic-algorithm-with-python-91fc701ff84), then it has been moved to the [TutorialProject](https://github.com/ahmedfgad/GARI/tree/master/TutorialProject) directory on 18 May 2020.* |
| 7 | + |
| 8 | +# PyGAD Installation |
| 9 | + |
| 10 | +To install [PyGAD](https://pypi.org/project/pygad), simply use pip to download and install the library from [PyPI](https://pypi.org/project/pygad) (Python Package Index). The library lives a PyPI at this page https://pypi.org/project/pygad. |
| 11 | + |
| 12 | +For Windows, issue the following command: |
| 13 | + |
| 14 | +```python |
| 15 | +pip install pygad |
| 16 | +``` |
| 17 | + |
| 18 | +For Linux and Mac, replace `pip` by use `pip3` because the library only supports Python 3. |
| 19 | + |
| 20 | +```python |
| 21 | +pip3 install pygad |
| 22 | +``` |
| 23 | + |
| 24 | +PyGAD is developed in Python 3.7.3 and depends on NumPy for creating and manipulating arrays and Matplotlib for creating figures. The exact NumPy version used in developing PyGAD is 1.16.4. For Matplotlib, the version is 3.1.0. |
| 25 | + |
| 26 | +# Project Steps |
| 27 | + |
| 28 | +The steps to follow in order to reproduce an image are as follows: |
| 29 | + |
| 30 | +- Read an image |
| 31 | +- Prepare the fitness function |
| 32 | +- Create an instance of the pygad.GA class with the appropriate parameters |
| 33 | +- Run PyGAD |
| 34 | +- Plot results |
| 35 | +- Calculate some statistics |
| 36 | + |
| 37 | +The next sections discusses the code of each of these steps. |
| 38 | + |
| 39 | +## Read an Image |
| 40 | + |
| 41 | +There is an image named `fruit.jpg` in the project which is read according to the next code. |
| 42 | + |
| 43 | +```python |
| 44 | +import imageio |
| 45 | +import numpy |
| 46 | + |
| 47 | +target_im = imageio.imread('fruit.jpg') |
| 48 | +target_im = numpy.asarray(target_im/255, dtype=numpy.float) |
| 49 | +``` |
| 50 | + |
| 51 | +Here is the read image. |
| 52 | + |
| 53 | + |
| 54 | + |
| 55 | +Based on the chromosome representation used in the example, the pixel values can be either in the 0-255, 0-1, or any other ranges. |
| 56 | + |
| 57 | +Note that the range of pixel values affect other parameters like the range from which the random values are selected during mutation and also the range of the values used in the initial population. So, be consistent. |
| 58 | + |
| 59 | +## Prepare the Fitness Function |
| 60 | + |
| 61 | +The next code creates a function that will be used as a fitness function for calculating the fitness value for each solution in the population. This function must be a maximization function that accepts 2 parameters representing a solution and its index. It returns a value representing the fitness value. |
| 62 | + |
| 63 | +The fitness value is calculated using the sum of absolute difference between genes values in the original and reproduced chromosomes. The `gari.img2chromosome()` function is called before the fitness function to represent the image as a vector because the genetic algorithm can work with 1D chromosomes. |
| 64 | + |
| 65 | +For more information about preparing the fitness function in PyGAD, please read the [PyGAD's documentation](https://pygad.readthedocs.io). |
| 66 | + |
| 67 | +```python |
| 68 | +target_chromosome = gari.img2chromosome(target_im) |
| 69 | + |
| 70 | +def fitness_fun(solution, solution_idx): |
| 71 | + fitness = numpy.sum(numpy.abs(target_chromosome-solution)) |
| 72 | + |
| 73 | + # Negating the fitness value to make it increasing rather than decreasing. |
| 74 | + fitness = numpy.sum(target_chromosome) - fitness |
| 75 | + return fitness |
| 76 | +``` |
| 77 | + |
| 78 | +## Create an Instance of the `pygad.GA` Class |
| 79 | + |
| 80 | +It is very important to use random mutation and set the `mutation_by_replacement` to `True`. Based on the range of pixel values, the values assigned to the `init_range_low`, `init_range_high`, `random_mutation_min_val`, and `random_mutation_max_val` parameters should be changed. |
| 81 | + |
| 82 | +If the image pixel values range from 0 to 255, then set `init_range_low` and `random_mutation_min_val` to 0 as they are but change `init_range_high` and `random_mutation_max_val` to 255. |
| 83 | + |
| 84 | +Feel free to change the other parameters or add other parameters. Please check the [PyGAD's documentation](https://pygad.readthedocs.io) for the full list of parameters. |
| 85 | + |
| 86 | +```python |
| 87 | +import pygad |
| 88 | + |
| 89 | +ga_instance = pygad.GA(num_generations=20000, |
| 90 | + num_parents_mating=10, |
| 91 | + fitness_func=fitness_fun, |
| 92 | + sol_per_pop=20, |
| 93 | + num_genes=target_im.size, |
| 94 | + init_range_low=0.0, |
| 95 | + init_range_high=1.0, |
| 96 | + mutation_percent_genes=0.01, |
| 97 | + mutation_type="random", |
| 98 | + mutation_by_replacement=True, |
| 99 | + random_mutation_min_val=0.0, |
| 100 | + random_mutation_max_val=1.0) |
| 101 | +``` |
| 102 | + |
| 103 | +## Run PyGAD |
| 104 | + |
| 105 | +Simply, call the `run()` method to run PyGAD. |
| 106 | + |
| 107 | +```python |
| 108 | +ga_instance.run() |
| 109 | +``` |
| 110 | + |
| 111 | +## Plot Results |
| 112 | + |
| 113 | +After the `run()` method completes, the fitness values of all generations can be viewed in a plot using the `plot_result()` method. |
| 114 | + |
| 115 | +```python |
| 116 | +ga_instance.plot_result() |
| 117 | +``` |
| 118 | + |
| 119 | +Here is the plot after 20,000 generations. |
| 120 | + |
| 121 | +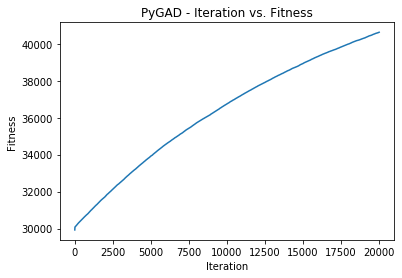 |
| 122 | + |
| 123 | +## Calculate Some Statistics |
| 124 | + |
| 125 | +Here is some information about the best solution. |
| 126 | + |
| 127 | +```python |
| 128 | +# Returning the details of the best solution. |
| 129 | +solution, solution_fitness, solution_idx = ga_instance.best_solution() |
| 130 | +print("Fitness value of the best solution = {solution_fitness}".format(solution_fitness=solution_fitness)) |
| 131 | +print("Index of the best solution : {solution_idx}".format(solution_idx=solution_idx)) |
| 132 | + |
| 133 | +if ga_instance.best_solution_generation != -1: |
| 134 | + print("Best fitness value reached after {best_solution_generation} generations.".format(best_solution_generation=ga_instance.best_solution_generation)) |
| 135 | + |
| 136 | +result = gari.chromosome2img(solution, target_im.shape) |
| 137 | +matplotlib.pyplot.imshow(result) |
| 138 | +matplotlib.pyplot.title("PyGAD & GARI for Reproducing Images") |
| 139 | +matplotlib.pyplot.show() |
| 140 | +``` |
| 141 | + |
| 142 | +# Evolution by Generation |
| 143 | + |
| 144 | +The solution reached after the 20,000 generations is shown below. |
| 145 | + |
| 146 | +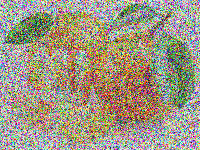 |
| 147 | + |
| 148 | +After more generations, the result can be enhanced like what shown below. |
| 149 | + |
| 150 | + |
| 151 | + |
| 152 | +The results can also be enhanced by changing the parameters passed to the constructor of the `pygad.GA` class. |
| 153 | + |
| 154 | +Here is an example of input image and how it is evolved after some iterations. |
| 155 | + |
| 156 | +<h1>Generation 0</h1> |
| 157 | + |
| 158 | + |
| 159 | + |
| 160 | +<h1>Generation 1,000</h1> |
| 161 | + |
| 162 | + |
| 163 | + |
| 164 | +<h1>Generation 2,500</h1> |
| 165 | + |
| 166 | + |
| 167 | + |
| 168 | +<h1>Generation 4,500</h1> |
| 169 | + |
| 170 | + |
| 171 | + |
| 172 | +<h1>Generation 7,000</h1> |
| 173 | + |
| 174 | + |
| 175 | + |
| 176 | +<h1>Generation 8,500</h1> |
| 177 | + |
| 178 | + |
| 179 | + |
| 180 | +<h1>Generation 20,000</h1> |
| 181 | + |
| 182 | +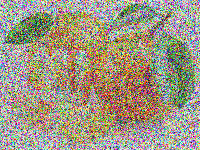 |
| 183 | + |
| 184 | +# For More Information |
| 185 | + |
| 186 | +There are different resources that can be used to get started with the building CNN and its Python implementation. |
| 187 | + |
| 188 | +## Tutorial: Reproduce Images with Genetic Algorithm |
| 189 | + |
| 190 | +In 1 May 2019, I wrote a tutorial discussing this project. The tutorial is titled [**Reproducing Images using a Genetic Algorithm with Python**](https://www.linkedin.com/pulse/reproducing-images-using-genetic-algorithm-python-ahmed-gad) which is published at Heartbeat. Check it at these links: |
| 191 | + |
| 192 | +- [Heartbeat](https://heartbeat.fritz.ai/reproducing-images-using-a-genetic-algorithm-with-python-91fc701ff84): https://heartbeat.fritz.ai/reproducing-images-using-a-genetic-algorithm-with-python-91fc701ff84 |
| 193 | +- [LinkedIn](https://www.linkedin.com/pulse/reproducing-images-using-genetic-algorithm-python-ahmed-gad): https://www.linkedin.com/pulse/reproducing-images-using-genetic-algorithm-python-ahmed-gad |
| 194 | + |
| 195 | +[](https://heartbeat.fritz.ai/reproducing-images-using-a-genetic-algorithm-with-python-91fc701ff84) |
| 196 | + |
| 197 | +## Book: Practical Computer Vision Applications Using Deep Learning with CNNs |
| 198 | + |
| 199 | +You can also check my book cited as [**Ahmed Fawzy Gad 'Practical Computer Vision Applications Using Deep Learning with CNNs'. Dec. 2018, Apress, 978-1-4842-4167-7**](https://www.amazon.com/Practical-Computer-Vision-Applications-Learning/dp/1484241665) which discusses neural networks, convolutional neural networks, deep learning, genetic algorithm, and more. |
| 200 | + |
| 201 | +Find the book at these links: |
| 202 | + |
| 203 | +- [Amazon](https://www.amazon.com/Practical-Computer-Vision-Applications-Learning/dp/1484241665) |
| 204 | +- [Springer](https://link.springer.com/book/10.1007/978-1-4842-4167-7) |
| 205 | +- [Apress](https://www.apress.com/gp/book/9781484241660) |
| 206 | +- [O'Reilly](https://www.oreilly.com/library/view/practical-computer-vision/9781484241677) |
| 207 | +- [Google Books](https://books.google.com.eg/books?id=xLd9DwAAQBAJ) |
| 208 | + |
| 209 | + |
| 210 | + |
| 211 | +# Contact Us |
| 212 | + |
| 213 | + |
| 214 | +* [LinkedIn](https://www.linkedin.com/in/ahmedfgad) |
| 215 | +* [Amazon Author Page](https://amazon.com/author/ahmedgad) |
| 216 | +* [Heartbeat](https://heartbeat.fritz.ai/@ahmedfgad) |
| 217 | +* [Paperspace](https://blog.paperspace.com/author/ahmed) |
| 218 | +* [KDnuggets](https://kdnuggets.com/author/ahmed-gad) |
| 219 | +* [TowardsDataScience](https://towardsdatascience.com/@ahmedfgad) |
| 220 | +* [GitHub](https://github.com/ahmedfgad) |
0 commit comments