diff --git a/README.md b/README.md
index 0446ed6..097c152 100644
--- a/README.md
+++ b/README.md
@@ -31,6 +31,14 @@ PDV supports visualizing N-linked intact glycopeptide identification result (ide
[ ](http://www.psidev.info/usi)
+
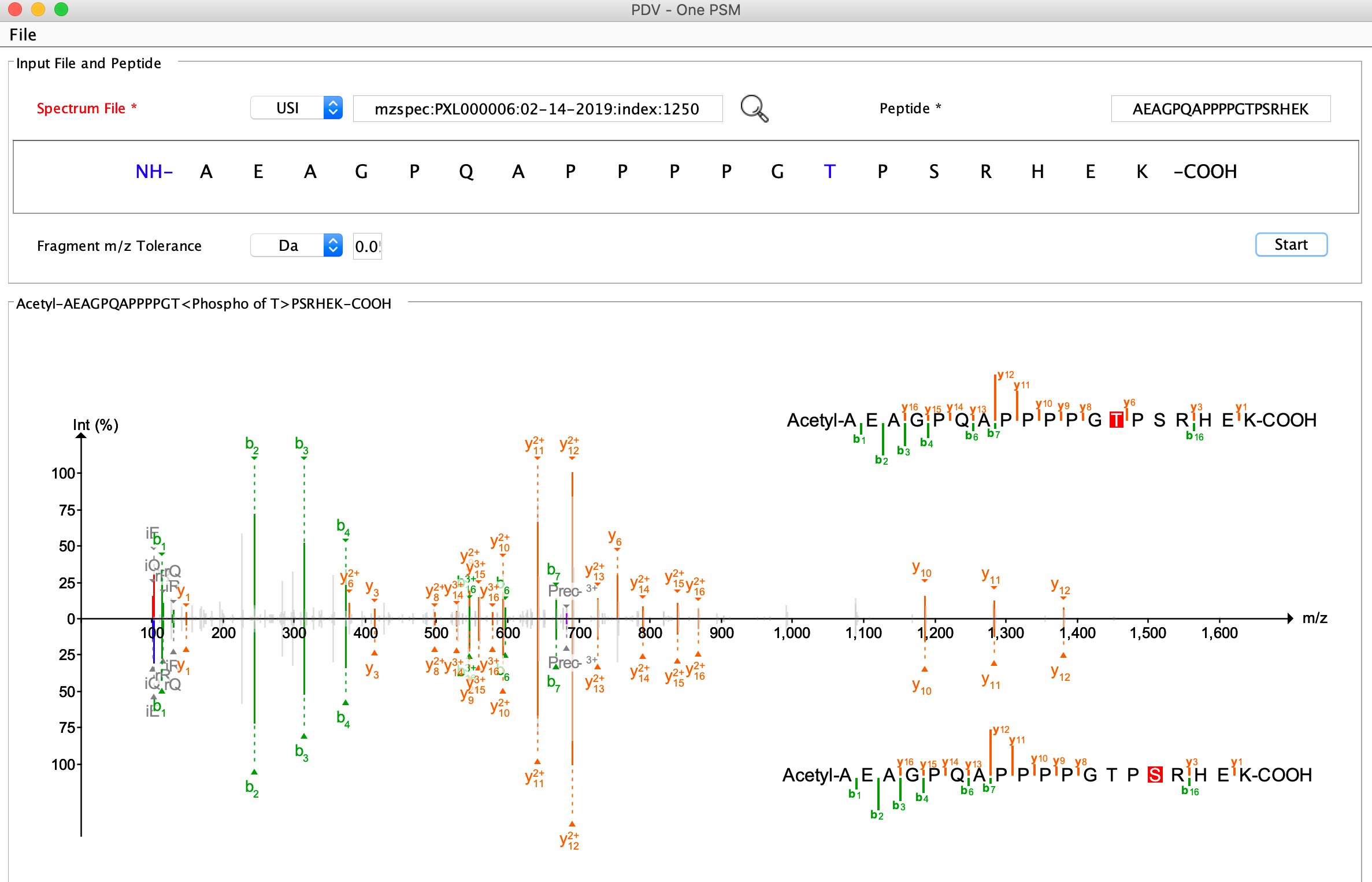
+#### [Mirror plot](https://github.com/wenbostar/PDV/wiki/Mirror-plot:-observed-vs-predicted-spectra) (Experimental spectrum VS predicted spectrum using deep learning)
+
+Top panel: experimental spectrum, bottom panel: predicted spectrum using deep learning.
+
+
](http://www.psidev.info/usi)
+
+#### [Mirror plot](https://github.com/wenbostar/PDV/wiki/Mirror-plot:-observed-vs-predicted-spectra) (Experimental spectrum VS predicted spectrum using deep learning)
+
+Top panel: experimental spectrum, bottom panel: predicted spectrum using deep learning.
+
+ +
+
#### Database searching:
| Software | Example files |
@@ -60,7 +68,7 @@ PDV supports visualizing N-linked intact glycopeptide identification result (ide
| Software | Example files |
| ----------------|:---------------|
-| [Casanovo](https://github.com/Noble-Lab/casanovo) | mgf:mzTab |
+| [Casanovo](https://github.com/Noble-Lab/casanovo) | [Manual](https://github.com/wenbostar/PDV/wiki/Visualize-Casanovo-result) |
| [Novor](https://www.ncbi.nlm.nih.gov/pubmed/26122521) | [mgf](http://pdv.zhang-lab.org/data/download/test_data/msdata/SF_200217_U2OS_TiO2_HCD_OT_rep1.mgf.gz):[csv](http://pdv.zhang-lab.org/data/download/test_data/novor/SF_200217_U2OS_TiO2_HCD_OT_rep1.novor.csv.gz) (only support the Novor result generated through [DeNovoGUI](https://github.com/compomics/denovogui)) |
| [DeepNovo](https://github.com/nh2tran/DeepNovo) | [mgf](http://pdv.zhang-lab.org/data/download/test_data/deepnovo/peaks.db.mgf.test.dup.mgf.gz):[txt](http://pdv.zhang-lab.org/data/download/test_data/deepnovo/output.deepnovo_db.tab) |
| [PepNovo+](http://proteomics.ucsd.edu/software-tools/531-2/) | [mgf](http://pdv.zhang-lab.org/data/download/test_data/msdata/SF_200217_U2OS_TiO2_HCD_OT_rep1.mgf.gz):[txt](http://pdv.zhang-lab.org/data/download/test_data/pepnovo/SF_200217_U2OS_TiO2_HCD_OT_rep1.mgf.out) |
+
+
#### Database searching:
| Software | Example files |
@@ -60,7 +68,7 @@ PDV supports visualizing N-linked intact glycopeptide identification result (ide
| Software | Example files |
| ----------------|:---------------|
-| [Casanovo](https://github.com/Noble-Lab/casanovo) | mgf:mzTab |
+| [Casanovo](https://github.com/Noble-Lab/casanovo) | [Manual](https://github.com/wenbostar/PDV/wiki/Visualize-Casanovo-result) |
| [Novor](https://www.ncbi.nlm.nih.gov/pubmed/26122521) | [mgf](http://pdv.zhang-lab.org/data/download/test_data/msdata/SF_200217_U2OS_TiO2_HCD_OT_rep1.mgf.gz):[csv](http://pdv.zhang-lab.org/data/download/test_data/novor/SF_200217_U2OS_TiO2_HCD_OT_rep1.novor.csv.gz) (only support the Novor result generated through [DeNovoGUI](https://github.com/compomics/denovogui)) |
| [DeepNovo](https://github.com/nh2tran/DeepNovo) | [mgf](http://pdv.zhang-lab.org/data/download/test_data/deepnovo/peaks.db.mgf.test.dup.mgf.gz):[txt](http://pdv.zhang-lab.org/data/download/test_data/deepnovo/output.deepnovo_db.tab) |
| [PepNovo+](http://proteomics.ucsd.edu/software-tools/531-2/) | [mgf](http://pdv.zhang-lab.org/data/download/test_data/msdata/SF_200217_U2OS_TiO2_HCD_OT_rep1.mgf.gz):[txt](http://pdv.zhang-lab.org/data/download/test_data/pepnovo/SF_200217_U2OS_TiO2_HCD_OT_rep1.mgf.out) |
 ](http://www.psidev.info/usi)
+
+#### [Mirror plot](https://github.com/wenbostar/PDV/wiki/Mirror-plot:-observed-vs-predicted-spectra) (Experimental spectrum VS predicted spectrum using deep learning)
+
+Top panel: experimental spectrum, bottom panel: predicted spectrum using deep learning.
+
+
](http://www.psidev.info/usi)
+
+#### [Mirror plot](https://github.com/wenbostar/PDV/wiki/Mirror-plot:-observed-vs-predicted-spectra) (Experimental spectrum VS predicted spectrum using deep learning)
+
+Top panel: experimental spectrum, bottom panel: predicted spectrum using deep learning.
+
+