The goal of this project is to enable you to utilize genomic big data in identifying regulatory mechanisms for differential expression (DE).
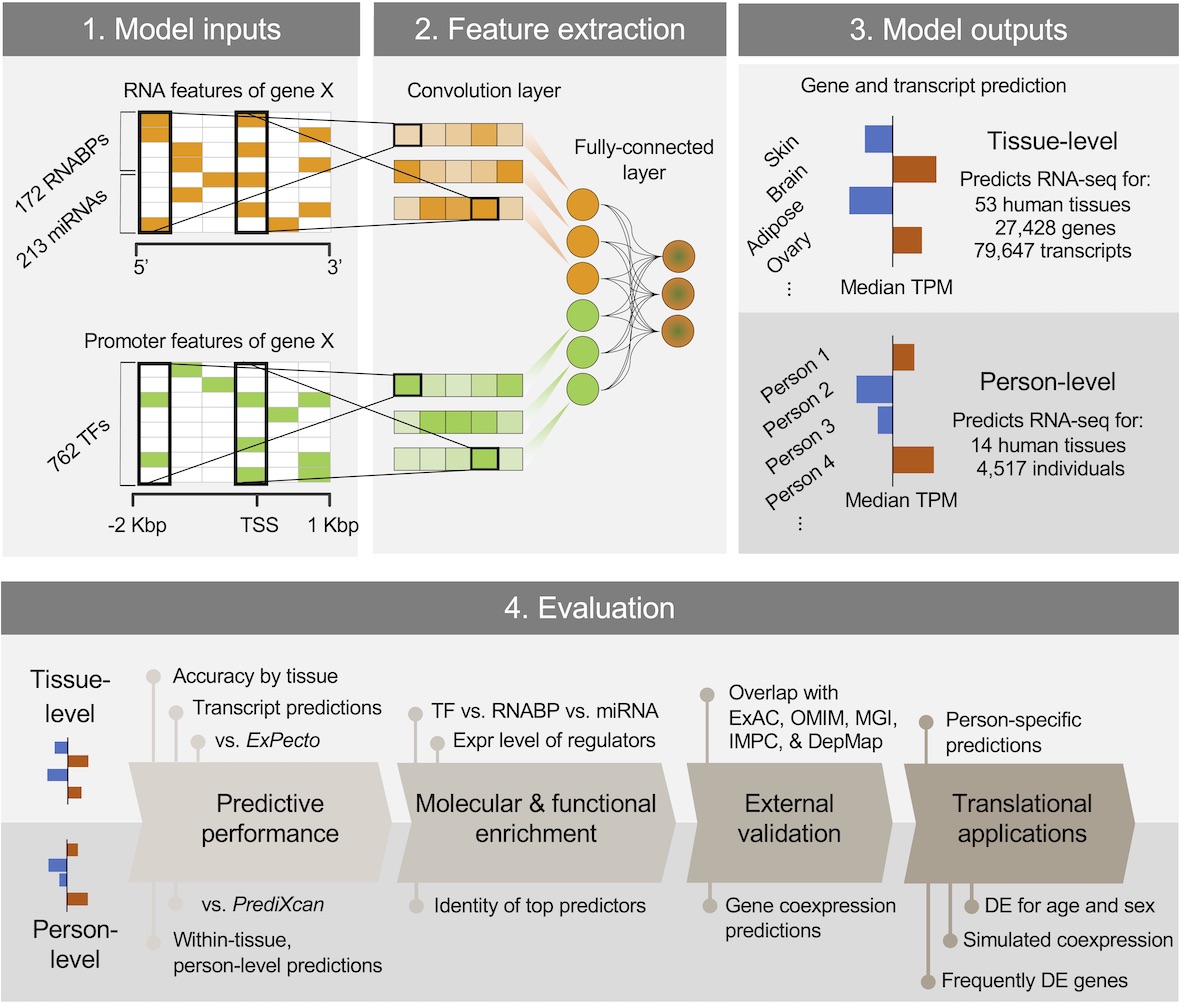
DEcode predicts inter-tissue variations and inter-person variations in gene expression levels from TF-promoter interactions, RNABP-mRNA interactions, and miRNA-mRNA interactions.
You can read more about this method in this paper (full text is available at https://rdcu.be/b5r3p) where we conducted a series of evaluation and applications by predicting transcript usage, drivers of aging DE, gene coexpression relationships on a genome-wide scale, and frequent DE in diverse conditions.
This tutorial shows you a way to run DEcode on Google Colab that provides you free access to a ready-to-use machine learning environment with a high-end GPU.
- Go to Google Colab and sign in to your Google account.
- Open Jupyter notebook.
- Menu -> File -> Open notebook -> GITHUB tab
- Search
https://github.com/stasaki/DEcode - Select
Run_DEcode_toy.ipynb
- Run each block of code.
Gene level features for all genes are available at data/GTEx53_gene/DEcode_data.tar.gz. However, due to the memory limitation in Google Colab, you cannot train a model with all genes. Please consider setting up Keras with GPU in your environment or use Code Ocean platform.
You can run DEcode on Code Ocean platform without setting up a computational environment. Our Code Ocean capsule provides reproducible workflows, all processed data, and pre-trained models for tissue- and person-specific transcriptomes and DEprior, at gene- or transcript level.
Tasaki, S., Gaiteri, C., Mostafavi, S. & Wang, Y. Deep learning decodes the principles of differential gene expression. Nature Machine Intelligence (2020) [link to paper] (full text is available at https://rdcu.be/b5r3p)
- GTEx transcriptome data - GTEx portal
- Transcription factor binding peaks - GTRD
- RNA binding protein binding peaks - POSTAR2
- miRNA binding locations - TargetScan